China has become a science powerhouse and it achieved this goal, in part, by sending its young scientitsts abroad to train in universities in Canada, Australia, United States, and Europe. Many of these countries have signed scientific cooperation agreements with China but some of those agreements are in danger of lapsing as China is increasingly seen as an untrustworthy enemy.
Read more »Category Archives: science
Posted by in science, Science Policy
Species-related publications
What’s a personal blog for, if not to blow my own horn? Well, it can only be to blow the horns of those who I…
The post Species-related publications appeared first on Evolving Thoughts.
Of Interest
One of the questions that have plagued my insomniac nights over the past decade or so is what makes something interesting. There are many proposals.…
The post Of Interest appeared first on Evolving Thoughts.
Posted by in Epistemology, Logic and philosophy, Philosophy, science, Theories
Do you understand the scientific literature?
I'm finding it increasingly difficult to understand the scientific literature even in subjects that I've been following for decades. Is it just because I'm getting too old to keep up?
Here's an example of a paper that I'd like to understand but after reading the abstract and the introduction I gave up. I'll quote the first paragraph of the introduction to see if any Sandwalk readers can do better.
I'm not talking about the paper being a complete mystery; I can figure out roughly what's it's about. What I'm thinking is that the opening paragraph could have been written in a way that makes the goals of the research much more comprehensible to average scientifically-literate people.
Weiner, D. J., Nadig, A., Jagadeesh, K. A., Dey, K. K., Neale, B. M., Robinson, E. B., ... & O’Connor, L. J. (2023) Polygenic architecture of rare coding variation across 394,783 exomes. Nature 614:492-499. [doi = 10.1038/s41586-022-05684-z]
Genome-wide association studies (GWAS) have identified thousands of common variants that are associated with common diseases and traits. Common variants have small effect sizes individually, but they combine to explain a large fraction of common disease heritability. More recently, sequencing studies have identified hundreds of genes containing rare coding variants, and these variants can have much larger effect sizes. However, it is unclear how much heritability rare variants explain in aggregate, or more generally, how common-variant and rare-variant architecture compare: whether they are equally polygenic; whether they implicate the same genes, cell types and genetically correlated risk factors; and whether rare variants will contribute meaningfully to population risk stratification.
The first question that comes to mind is whether the variant that's associated with a common disease is the cause of that disease or merely linked to the actual cause. In other words, are the associated variants responsible for the "effect size"? It sounds like the answer is "yes" in this case. Has that been firmly esablished in the GWAS field?
Birds of a feather: epigenetics and opposition to junk DNA
There's an old saying that birds of a feather flock together. It means that people with the same interests tend to associate with each other. It's extended meaning refers to the fact that people who believe in one thing (X) tend to also believe in another (Y). It usually means that X and Y are both questionable beliefs and it's not clear why they should be associated.
I've noticed an association between those who promote epigenetics far beyond it's reasonable limits and those who reject junk DNA in favor of a genome that's mostly functional. There's no obvious reason why these two beliefs should be associated with each other but they are. I assume it's related to the idea that both beliefs are presumed to be radical departures from the standard dogma so they reinforce the idea that the author is a revolutionary.
Or maybe it's just that sloppy thinking in one field means that sloppy thinking is the common thread.
Here's an example from Chapter 4 of a 2023 edition of the Handbook of Epigenetics (Third Edition).
The central dogma of life had clearly established the importance of the RNA molecule in the flow of genetic information. The understanding of transcription and translation processes further elucidated three distinct classes of RNA: mRNA, tRNA and rRNA. mRNA carries the information from DNA and gets translated to structural or functional proteins; hence, they are referred to as the coding RNA (RNA which codes for proteins). tRNA and rRNA help in the process of translation among other functions. A major part of the DNA, however, does not code for proteins and was previously referred to as junk DNA. The scientists started realizing the role of the junk DNA in the late 1990s and the ENCODE project, initiated in 2003, proved the significance of junk DNA beyond any doubt. Many RNA types are now known to be transcribed from DNA in the same way as mRNA, but unlike mRNA they do not get translated into any protein; hence, they are collectively referred to as noncoding RNA (ncRNA). The studies have revealed that up to 90% of the eukaryotic genome is transcribed but only 1%–2% of these transcripts code for proteins, the rest all are ncRNAs. The ncRNAs less than 200 nucleotides are called small noncoding RNAs and greater than 200 nucleotides are called long noncoding RNAs (lncRNAs).
In case you haven't been following my blog posts for the past 17 years, allow me to briefly summarize the flaws in that paragraph.
- The central dogma has nothing to do with whether most of our genome is junk
- There was never, ever, a time when knowledgeable scientists defended the idea that all noncoding DNA is junk
- ENCODE did not "prove the significance of junk DNA beyond any doubt"
- Not all transcripts are functional; most of them are junk RNA transcribed from junk DNA
So, I ask the same question that I've been asking for decades. How does this stuff get published?
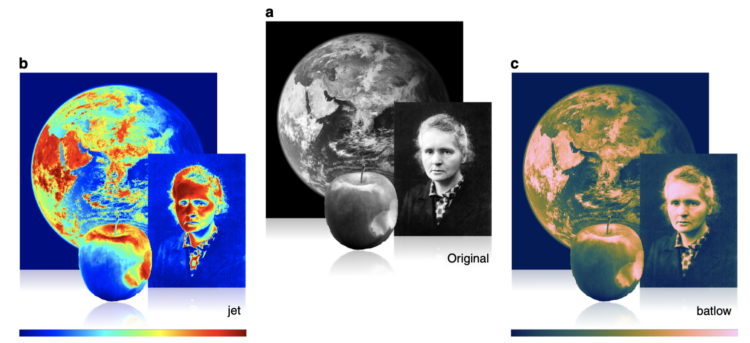
Misuse of the rainbow color scheme to visualize scientific data
Fabio Crameri, Grace Shephard, and Philip Heron in Nature discuss the drawbacks of using the rainbow color scheme to visualize data and more readable alternatives:
The accurate representation of data is essential in science communication. However, colour maps that visually distort data through uneven colour gradients or are unreadable to those with colour-vision deficiency remain prevalent in science. These include, but are not limited to, rainbow-like and red–green colour maps. Here, we present a simple guide for the scientific use of colour. We show how scientifically derived colour maps report true data variations, reduce complexity, and are accessible for people with colour-vision deficiencies. We highlight ways for the scientific community to identify and prevent the misuse of colour in science, and call for a proactive step away from colour misuse among the community, publishers, and the press.
Jupiter weighs two quettagrams
New names for very large and very small weights and sizes have been adopted.
Last November's meeting of the General Conference on Weights and Measures wasn't covered by the major media outlets so you probably don't know that an electron now weighs one rontogram and the diameter of the universe is about one ronnameter [SI units get new prefixes for huge and tiny numbers].
The official SI prefixes for very large things are now ronna (1027) and quetta (1030) and the prefixes for very small things are ronto (10-27) and quecto (10-30).
This is annoying because we've just gotten used to zetta, yotta, zepto, and yocto (adopted in 1991). I suspect that the change was prompted by the huge storage capacity of your latest smartphone (several yottabytes) and the wealth of the world's richest people (several zeptocents). Or maybe it was the price of houses in Toronto. Or something like that. In any case, we needed to prepare for kilo or mega increases.
The bad news is that the latest additions used up the last two available letters of the alphabet so if things get any bigger or smaller we may have to add a few more letters to the alphabet.
Posted by in science, Science Education