Category Archives: Workshop
Announcing NCBI’s First Ever BioEd Summit!
Posted by in education, What's New, Workshop
Celebrating 1 Year of NCBI Virtual Outreach Events
Posted by in codeathon, education, NCBI Education, training, webinars, What's New, Workshop
NLM announces rescheduled Curation at Scale Workshop
Data curation plays a critical role in today’s biomedical research and ensures scientific data will be accessible for future research and reuse. In the time of pandemics, the need to get scientific information to researchers, medical personnel, and the public as quickly as possible is greater than ever before. In response to the need for … Continue reading NLM announces rescheduled Curation at Scale Workshop
The post NLM announces rescheduled Curation at Scale Workshop appeared first on NCBI Insights.
Posted by in conferences, Data curation, data science, Genome, What's New, Workshop
Distinguishability in Phylogenetic Networks, report
We have now completed the workshop, as you can tell from the previous post with some photos. Here is a brief report on what seem to me to be some of the more useful points covered.
We had 10 formal presentations, but we also focused on group discussions for several hours each day. It may be the latter that were the most productive. However, I will briefly summarize the talks first.
I spent my time time in the opening talk emphasizing the different viewpoints of network computations, which focus on the patterns that can be detected in the data, and the network users, who are generally more interested in the processes that create those patterns (or are, indeed, absence from the patterns but present in the phylogenetic history, anyway). This highlights the two essential point of the workshop title, that both the patterns and the processes are much harder to untangle for networks than for trees.
Céline Scornavacca then bravely tried to tackle the combined problem, anyway, by trying to produce networks from analyzing the patterns in terms of their processes. The issues immediately become obvious, but she seems to be determined to proceed, regardless. Later in the week, Luay Nakhleh reduced the issue simply to vertical processes (including incomplete lineage sorting but not gene duplication-loss) versus horizontal processes. This creates a tractable problem for parsimony and likelihood, but the current challenge remains the limited number of taxa.
Vincent Moulton, Cécile Ané and Charles Semple dodged the issue by focusing on computations. Charles took on the challenge of trying to create a network version of Neighbor-Joining, which would address the issues of computational speed and taxon sampling, and Vince tackled super-networks, and the conditions required for building networks from a collection of smaller (ie. incomplete) trees. Both topics remain open questions. Cécile, on the other hand, discussed network models for trait evolution, which is important for the use of phylogenetic comparative methods when using networks.
On the user side, the presentations focused on examples, and the issues encountered when dealing with them. James Whitfield and Axel Janke talking about biology (mostly phylogenomics), while Johann-Mattis List talked about linguistics, and Tiago Tresoldi talked about stemmatology. In some ways, historical linguistics seems to be the odd one out, since many of the issues dealt with are somewhat removed from those in the other fields. However, in biology there are actually two options for producing networks — directly from the data or via "gene trees" (trees derived from non-recombining blocks of sequences). For the humanities, much of the current discussion is about the nature of the data, and how to code it for quantitative analysis.
This brings us to the discussions. While some time was spent on trying to establish whether biologists think that there is a difference between lateral gene transfer and horizontal gene transfer, or between incomplete lineage sorting, ancestral polymorphism and deep coalescence, some productive interchanges also occurred. Here is a coverage of four of the most important ones.
There was general agreement that there are several barriers to widespread adoption of network analyses in phylogenetics. This includes the development of suitable methods (in the face on indistinguishability), but also includes an understanding of what methods are currently available, what data are required to apply those methods, what taxon sampling is required to benefit from the methods, and how to use the programs that implement those methods.
One popular suggestion was therefore to produce some sort of "cookbook", to address the complexity of producing networks, given that there are many methods and programs. From the users' point of view this would illustrate what network analyses can do, in terms of finding reticulation patterns in the data; and from the computational point of view it would outline what needs to be done to get the programs to work. The consensus idea was to choose two suitable datasets (yet to be determined), and then have each program author provide analyses of them (including any scripts that are needed).
Following on from this latter point, it was agreed that the programs need easy user interfaces, if they are to become more widely used. Here, the word "widely" includes casual users from outside of phylogenetics, who use phylogenies as only one of many tools in their work. So, users will include those who need nothing more than a "point and click" control panel (which may be >90% of potential users) to those who would benefit from scripting control of the analyses. The interface needs both a front end, to specify the particular analysis, and a back end, to allow exploration of the output.
Another long-discussed issue was how to popularize networks, which is clearly a major topic. A phylogenetic tree is nothing more than one of the possible networks for any given dataset, and yet the focus is often on trees rather than networks.
To this end, it was noted that the current Wikipedia entry is inadequate, especially compared to the corresponding entry for phylogenetic trees. Not only is this entry out of date, it is in a number of ways misleading. In particular, there needs to be a discussion of the fact that, if a network is a "tree with reticulations", then ignoring the reticulations can result in the wrong tree, and the branch lengths may be severely under-estimated. There are challenges to getting Wikipedia entries changed, especially the wholesale re-writing of an entry, but this will be necessary.
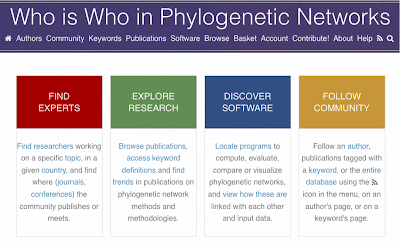
Finally, it was noted that Philippe Gambette's Who is Who in Phylogenetic Networks website is extremely useful but is still poorly known, even within the phylogenetic networks community. We had a long discussion about how to enhance this site, to make it a more general-purpose repository of information about phylogenetic networks. This included a more inclusive database, more comprehensive tagging of keywords, enhanced descriptions of those keywords, and ways to keep the database up to date.
Steven Kelk has the notes from the final session, which was a review of what we achieved during the workshop, and which contains the To Do list. Both he and Philippe have the notes about modifications for the Who is Who in Phylogenetic Networks website, which is likely to be the first outcome-project tackled.
Thankyou to everybody who participated in the workshop. It seemed to be very productive, with a number of concrete outcomes that will be interesting to review at the next workshop.
Distinguishability in Phylogenetic Networks, photos
Evidence that we were in the Netherlands.
Evidence that we did some work.
Left to right: Steven Kelk, David Morrison, Mike Steel, Philippe Gambette (obscured), Tiago Tresoldi, Claudia Solis-Lemus, Fabio Pardi, Simone Linz, Mark Jones.
Left to right: David Morrison, Cecile Ané, Philippe Gambette (obscured), Katharina Huber, Leen Stougie, Remie Janssen, Yukihiro Murakami, Mattis List, Gereon Kaiping and Charles Semple.
Left to right: David Morrison (obscured), Axel Janke, Steven Kelk, Charles Semple, Claudia Solis-Lemus, Mark Jones (obscured), Fabio Pardi, Leo van Iersel, Simone Linz and Vincent Moulton.
Céline Scornavacca lectures Cecile Ané.
Axel Janke and Leo van Iersel contemplate methods for infering hybridization.
Philippe Gambette and Guido Grimm.
Mozes Blom and Jim Whitfield.
Mike Steel and Luay Nakhleh.
Luay delivers his Final Message, to Mozes Blom, Cecile Ané, Katharina Huber and Charles Semple.
Capturing phylogenetic algorithms for linguistics
A little over a week ago I was at a workshop "Capturing phylogenetic algorithms for linguistics" at the Lorentz Centre in Leiden (NL). This is, as some of you will recall, the venue that hosted two earlier workshops on phylogenetic networks in 2012 and 2014.
I had been invited to participate and to give a talk and I chose to discuss the possible relevance of phylogenetic networks (as opposed to phylogenetic trees) for linguistics. (My talk is here). This turned out to be a good choice because, although phylogenetic trees are now a firmly established part of contemporary linguistics, networks are much less prominent. Data-display networks (which visualize incongruence in a data-set, but do not model the genealogical processs that gave rise to it) have found their way into some linguistic publications, and a number of the presentations earlier in the week showed various flavours of split networks. However, the idea of constructing "evolutionary" phylogenetic networks - e.g. modeling linguistic analogues of horizontal gene transfer - has not yet gained much traction in the field. In many senses this is not surprising, since tools for constructing evolutionary phylogenetic networks in biology are not yet widely used, either. As in biology, much of the reticence concerning these tools stems from uncertainty about whether models for reticulate evolution are sufficiently mature to be used 'out of the box'.
As far as this blog is concerned the relevant word in linguistics is 'borrowing'. My lay-man interpretation of this is that it denotes the process whereby words or terms are transferred horizontally from one language to another. (Mattis, feel free to correct me...) There were many discussions of how this proces can confound the inference of concept and language trees, but other than it being a problem there was not a lot a said about how to deal with it methodologically (or model it). One of the issues, I think, is that linguists are nervous about the interface between micro and macro levels of evolution and at what scale of (language) evolution horizontal events could and should be modelled. To cite a biological analogue: if you study populations at the most microscopic level evolution is usually reticulate (because of e.g. meiotic recombination) but at the macro level large parts of mammalian evolution are uncontroversially tree-like. In this sense whether reticulate events are modeled depends on the event itself and the scale of the phylogenetic model concerned.
Are there analogues of population-genetic phenomena in linguistics, and are they foundations for phenomena observed at the macro level? Is there a risk of over-stating the parallels with biology? One participant told me that, while she felt that there was definitely scope for incrorporating analogies of species and gene trees within linguistics - and many of the participants immediately recognized these concepts - comparisons quickly break down at more micro levels of evolution.
I'm not the right person to comment on this of course, or to answer these questions, but in any case it's clear that linguistics has plenty of scope for continuing the horizontal/vertical discussions that have already been with us for many years in biology...
Last, but not least: it was a very enjoyable workshop and I'm grateful to the organizers for inviting me!
Posted by in Evolutionary network, Linguistics, Workshop
Capturing phylogenetic algorithms for linguistics
A little over a week ago I was at a workshop "Capturing phylogenetic algorithms for linguistics" at the Lorentz Centre in Leiden (NL). This is, as some of you will recall, the venue that hosted two earlier workshops on phylogenetic networks in 2012 and 2014.
I had been invited to participate and to give a talk and I chose to discuss the possible relevance of phylogenetic networks (as opposed to phylogenetic trees) for linguistics. (My talk is here). This turned out to be a good choice because, although phylogenetic trees are now a firmly established part of contemporary linguistics, networks are much less prominent. Data-display networks (which visualize incongruence in a data-set, but do not model the genealogical processs that gave rise to it) have found their way into some linguistic publications, and a number of the presentations earlier in the week showed various flavours of split networks. However, the idea of constructing "evolutionary" phylogenetic networks - e.g. modeling linguistic analogues of horizontal gene transfer - has not yet gained much traction in the field. In many senses this is not surprising, since tools for constructing evolutionary phylogenetic networks in biology are not yet widely used, either. As in biology, much of the reticence concerning these tools stems from uncertainty about whether models for reticulate evolution are sufficiently mature to be used 'out of the box'.
As far as this blog is concerned the relevant word in linguistics is 'borrowing'. My lay-man interpretation of this is that it denotes the process whereby words or terms are transferred horizontally from one language to another. (Mattis, feel free to correct me...) There were many discussions of how this proces can confound the inference of concept and language trees, but other than it being a problem there was not a lot a said about how to deal with it methodologically (or model it). One of the issues, I think, is that linguists are nervous about the interface between micro and macro levels of evolution and at what scale of (language) evolution horizontal events could and should be modelled. To cite a biological analogue: if you study populations at the most microscopic level evolution is usually reticulate (because of e.g. meiotic recombination) but at the macro level large parts of mammalian evolution are uncontroversially tree-like. In this sense whether reticulate events are modeled depends on the event itself and the scale of the phylogenetic model concerned.
Are there analogues of population-genetic phenomena in linguistics, and are they foundations for phenomena observed at the macro level? Is there a risk of over-stating the parallels with biology? One participant told me that, while she felt that there was definitely scope for incrorporating analogies of species and gene trees within linguistics - and many of the participants immediately recognized these concepts - comparisons quickly break down at more micro levels of evolution.
I'm not the right person to comment on this of course, or to answer these questions, but in any case it's clear that linguistics has plenty of scope for continuing the horizontal/vertical discussions that have already been with us for many years in biology...
Last, but not least: it was a very enjoyable workshop and I'm grateful to the organizers for inviting me!
Posted by in Evolutionary network, Linguistics, Workshop
Singapore, Day 5
Louxin Zhang started with a couple of proofs about what he called "stable" networks; and Stefan Grünewald developed his thoughts on quartet algoritms for splits graphs. At the other extreme, Nadine Ziemert talked entirely biology, introducing the audience to the problem of trying to study the evolution of secondary metabolites. In between, Eric Tannier tried to use horizontal gene transfer to date the nodes of networks, assuming that HGT requires a temporally consistent network. Francois-Joseph Lapointe produced the only really statistical talk of the week, trying to produce p-values for patterns on sequence similarity networks.
Daniel Huson popped in for the last day, and presented us with some ideas for the future development of both SplitsTree (unrooted networks) and Dendroscope (rooted networks). Apparently, the need is for SplitsTree to handle larger sets of trees, while for Dendroscope it is to produce networks from pairs of input trees. He also noted that there are still more networks being produced using median joining rather than neighbor-net, due to the amount of work being done on human mitochondrial sequences.
An interest was expressed in continuing the series of meetings on phylogenetic networks (Leiden 2012, Leiden 2014) — I first met most of the people working on networks in phylogenetics in Uppsala in 2004 (Phylogenetic Combinatorics and Applications).
Today we also celebrated Dan Gusfield's 2^6 birthday, with a strawberry cream cake.
So, all in all, a very successful meeting.
After the sessions finished, I went down to the Gardens By The Bay to look at the Supertree Grove. As you can see, a "super" tree is by any definition actually a network.
Singapore, Day 4
There was more heavy maths today.
Charles Semple started by counting trees within specified types of network. In the process, he provided the first mathematical proof of the week (he actually provided two). He also raised the issue of what, exactly, is a phylogenetic network — we have had many mathematical restrictions placed on networks this week, and it is not always clear how any of them might relate to biological concepts.
Leo van Iersel tried constructing super-networks from incomplete sub-networks, sticking to algorithms rather than proofs. Yufeng Wu and Zhi-Zhong Chen later tried the same strategy for their networks, as did Lusheng Wang for pedigree comparison (he was the only person other than myself to even mention pedigrees).
Mike Steel considered under what circumstances a network can be viewed as a "tree with reticulations" rather than a non-tree network (ie. not every vertex is part of the same underlying tree); this led him to the interesting observation that whether a dataset can be represented by a tree can depend on the taxon sampling. He also looked at when a set of non-tree distances can appear to be tree-like, which is the sort of question that only a mathematician would ask.
Most of the audience interjections this week have come from Sagi Snir, and the rest of the speakers got to return the favor this afternoon, when he spoke about trying to reconstruct trees subject to large amounts of horizontal gene transfer. In the process, he also tried to "sketch" a mathematical proof, which turned into a full-sized painting, before moving on to his algorithm.
Singapore, Day 3
It was hard going this morning for the biologists, as there were three main computational talks. First, Vince Moulton further developed some of his ideas about split networks, including median networks, quasi-median networks and neighbor-nets, and what sorts of trees they might contain. Then, Céline Scornavacca expanded on her ideas for calculating the "hybrid number". Finally, Jens Lagergren outlined his work on fitting gene trees to known species trees and networks; this has come a long way in recent years.
We had the afternoon off, although many people took the opportunity to pretend that they were still in their offices at home. Myself, I sat by the pool waiting for the temperature to cool (this was the hottest day so far this week), and then went to the Singapore Botanical Gardens, where I circumnavigated the Evolution Garden, the National Orchid Garden, and the Rainforest Walk. I then briefly perused Orchard Street (one of the most ridiculous shopping meccas you will ever see) and the Raffles Hotel (an even more ridiculous hang-over from British imperialism), before returning to the pool side. It's a tough life.