Over 60 years ago, Robert Sokal and Peter Sneath changed the way we quantitatively study evolution, by providing the first numerical approach to infer a phylogenetic tree. About the same time, but in German, Willi Hennig established the importance of distinguishing primitive and advanced character states, rather than treating all states as equal. This established a distinction between phenetics and phylogenetics; and the latter is the basis of all modern studies, whether it is explicitly acknowledged or not.
More than two decades later, Steve Farris and the Willi Hennig Society (WHS) established parsimony as the standard approach for evaluating character-state changes for tree inference. In this approach, morphological traits are scored and arranged data matrices, and then the most parsimonious solution is found to explain the data. This tree, usually a collection of most-parsimonious trees (MPT), was considered to be the best approximation of the true tree. Clades in the trees were synonymized with monophyly sensu Hennig (1950, short English version published 1965), and grades with paraphyly: Cladistics was born (see also: Let's distinguish between Hennig and Cladistics).
Why parsimony? Joe Felsenstein, who was not a member of the WHS but brought us, among many other things, the nonparametic bootstrap (Felsenstein 1985), put it like this (Felsenstein 2001):
History: William of Ockham told Popper to tell Hennig to use parsimonySoon, parsimony and cladistics came under threat by advances in computer technology and Kary Mullis' development of the polymerase-chain-reaction (PCR; Mullis & Faloona 1987) in the early 80s (note: Mullis soon went on with more fun stuff, outside science). While the data analysis took ages (literally) in the early days, more and more speedy heuristics were invented for probabilistic inferences. PCR marked the beginning of the Molecular Revolution, and genetic data became easy to access. Soon, many researchers realized that parsimony trees perform badly for this new kind of data, a notion bitterly rejected by the parsimonists, organized mainly in the WHS: the "Phylogenetic Wars" raged.
The parsimony faction lost. Today, when we analyze (up to) terabytes of molecular data, we use probabilistic methods such as maximum likelihood (ML) and Bayesian inference (BI). However, one parsimony bastion has largely remained unfazed: palaeontology.
In a series of new posts, we will try to change that; and outline what easy-to-compute networks have to offer when analyzing non-molecular data.
It's just similarity, stupid!
One collateral damage of the Phylogenetic Wars was distance-based methods, which, still today, are sometimes classified as "phenetic" in opposite to the "phylogenetic" character-based methods (parsimony, ML, BI). The first numerical phylogenetic trees were not based on character matrices but distance matrices (eg. Michener & Sokal 1957 using a cluster algorithm; Cavalli-Sforza & Edwards 1965 using parsimony; see also Felsenstein 2004, pp.123ff).
But no matter which method, optimality criterion or data-type we use, in principal we do the analysis under the same basic assumptions:
- the more closely related two taxa are, then the more similar they should be.
- the more similar two taxa are, then the more recent is their split.
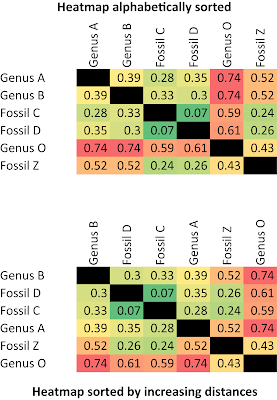
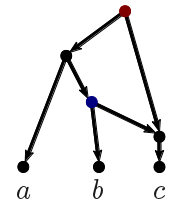
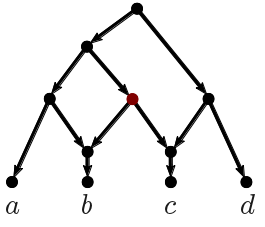
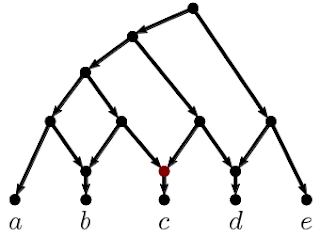
The ingroup (blue clade) and outgroup (red clade) are most distant from each other. Placing the fossils is trivial: Z is closer to O than to C, the member of the ingroup with the fewest advanced character states. Ingroup sister taxa A + C and B + D are most similar to each other. The monophyly of the ingroup and its two subclades is perfectly reflected by the inter-taxon distances.
Assuming that the character distance reflects the phylogenetic distance (ie. the distance along the branches of the true tree), any numerical phylogenetic approach will succeed in finding the true tree. The Neighbor-Joining method (using either the Least Squares or Minimum Evolution criteria) will be the quickest calculation. The signal from such matrices is trivial, we are in the so-called "Farris Zone" (defined below).
We wouldn't even have to infer a tree to get it right (ie. nested monophyly of A + C, B + D, A–D), we could just look at the heat map sorted by inter-taxon distance.
Just from the distance distributions, visualized in the form of a "heat-map", it is obvious that A–D are monophyletic, and fossil Z is part of the outgroup lineage. As expected for the same phylogenetic lineage (because changes accumulate over time), its fossils C and D are still relatively close, having few advanced character states, while the modern-day members A and B are have diverged from each other (based on derived character states). Taxon B is most similar to D, while C is most similar A. So, we can hypothesize that C is either a sister or precursor of A, and D is the same of B. If C and D are stem group taxa (ie. they are paraphyletic), then we would expect that both would show similar distances to A vs. B, and be closer to the outgroup. If representing an extinct sister lineage (ie. CD is monophyletic), they should be more similar to each other than to A or B. In both cases (CD either paraphyletic or monophyletic), A and B would be monophyletic, and so they should be relatively more similar to each other than to the fossils as well.
Having a black hole named after you
The Farris Zone is that part of the tree-space where the signals from the data are trivial, we have no branching artifacts, and any inference (tree or network), gives us the true tree.
It's opposite has been, unsurprisingly perhaps, labeled the "Felsenstein Zone". This is the part of the tree-space where branching artifacts are important — the inferred tree deviates from the true tree. Clades and grades (structural aspects of the tree) are no longer synonymous with monophyly and paraphyly (their evolutionary interpretation).
We can easily shift our example from the Farris into the Felsenstein Zone, by halving the distances between the fossils and the first (FCA) and last (LCA) common ancestors of ingroup and outgroup and adding some (random = convergence; lineage-restricted = homoiology) homoplasy to the long branches leading to the modern-day genera.
The difference between distance, parsimony and probabilistic methods is how we evaluate alternative tree topologies when similarity patterns become ambiguous — ie. when we approach or enter the Felsenstein Zone. Have all inferred mutations the same probability; how clock-like is evolution; are their convergence/ saturation effects; how do we deal with missing data?
For our example, any tree inference method will infer a wrong AB clade, because the fossils lack enough traits shared only with their sisters but not with the other part of the ingroup. Only the roots are supported by exclusively shared (unique) derived traits (Hennigian "synapomorphies"). The long-branch attraction (LBA) between A and B is effectively caused by:
- 'short-branch culling' between C and D: the fossils are too similar to each other; and their modern relatives too modified;
- the character similarity between A and B underestimates the phylogenetic distance between A and B, due to derived traits that evolved in parallel (homoiologies).
We have no ingroup-outgroup LBA, because the three convergent traits shared by O and A or B, respectively, compete with a total of eight lineage-unique and conserved traits (synapomorphies) — six characters are compatible with a O-A or O-B sister-relationship (clade in a rooted tree) but eight are incompatible. We correctly infer an A–D | O + Z split (ie. A–D clade when rooted with O) simply because A and B are still more similar to C and D than to Z and O; not some method- or model-inflicted magic.
The magic of non-parametric bootstrapping
When phylogeneticists perform bootstrapping, they usually do it to try to evaluate branch support values — a clade alone is hardly sufficient to infer an inclusive common origin (Hennig's monophyly), so we add branch support to quantify its quality (Some things you probably don't know about the bootstrap). In palaeontology, however, this is not a general standard (Ockhams Razor applied but not used), for one simple reason: bootstrapping values for critical branches in the trees are usually much lower than the molecular-based (generally accepted) threshold of 70+ for "good support" (All solved a decade ago).
When we bootstrap the Felsenstein Zone matrix that gives us the "wrong" (paraphyletic) AB clade, no matter which tree-inference method we use, we can see why this standard approach undervalues the potential of bootstrapping to explore the signal in our matrices.
While parsimony and NJ bootstrap pseudoreplicates either fall prey to LBA or don't provide any viable alternative (the bootstrap replicate matrix lacks critical characters), in the example a significant amount of ML pseudoreplicates did escape the A/B long-branch attraction.
Uncorrected, the correct splits A + C vs. rest and B + D vs. rest can be found in 19% of the 10,000 computed pseudoreplicate trees. When correcting for ascertainment bias, their number increases to 41%, while the support for the wrong A + B "clade" collapses to BSML = 49. Our BS supports are quite close to what Felsenstein writes in his 2004 book: For the four taxon case, ML has a 50:50 chance to escape LBA (BSML = true: 41 vs. false: 49), while MP and distance-methods will get it always wrong (BSMP = 88, BSNJ = 86).
The inferred tree may get it wrong but the (ML) bootstrap samples tell us the matrix' signal is far from clear.
Side-note: Bayesian Inference cannot escape such signal-inherent artifacts because its purpose is to find the tree that best matches all signals in the data, which, in our case, is the wrong alternative with the AB clade — supported by five characters, rejected by four each including three that are largely incompatible with the true tree. Posterior Probabilities will quickly converge to 1.0 for all branches, good and bad ones (see also Zander 2004); unless there is very little discriminating signal in the matrix — a CD clade, C sister to ABD, D sister to ABC, ie. the topological alternatives not conflicting with the wrong AB clade, will have PP << 1.0 because these topological alternatives give similar likelihoods.
Long-edge attraction
When it comes to LBA artifacts and the Felsenstein Zone, our preferred basic network-inference method, the Neighbor-Net (NNet), has its limitations, too. The NNet algorithm is in principle a 2-dimensional extension of the NJ algorithm. The latter is prone to LBA, hence, and the NNet can be affected by LEA: long edge attraction. The relative dissimilarity of A and B to C and D, and (relative) similarity of A/B and C/D, respectively, will be expressed in the form of a network neighborhood.
Note, however, the absence of a C/D neighbourhood. If C is a sister of D (as seen in the NJ tree), then there should be at least a small neighbourhood. It's missing because C has a different second-best neighbour than B within the A–D neighbourhood. While the tree forces us into a sequence of dichotomies, the network visualizes the two competing differentiation patterns: general advancement on the one hand (ABO | CDZ split), and on the other potential LBA/LEA, vs. similarity due to a shared common ancestry (ABCD | OZ split; BD neighborhood).
Just from the network, we would conclude that C and D are primitive relatives of A and B, potentially precursors. The same could be inferred from the trees; but if we map the character changes onto the net (Why we may want to map trait evolution on networks), we can notice there may be more to it.
Future posts
In each of the upcoming posts in this (irregular) series, we will look at a specific problem with non-molecular data, and test to what end exploratory data analysis can save us from misleading clades; eg. clades in morphology-informed (parts of, in case of total evidence) trees that are not monophyletic.
* The uncorrected ML tree shows branch lengths that are unrealistic (note the scale), and highly distorted. The reason for this is that the taxon set includes (very) primitive fossils and (highly) derived modern-day genera, but the matrix has no invariable sites telling the ML optimization that changing from 0 ↔ 1 is not that big of a deal. This is where the ascertainment bias correction(s) step(s) in (RAxML-NG, the replacement for classic RAxML 8 used here, has more than one implementation to correct for ascertainment bias. A tip for programmers and coders: effects of corrections have so far not been evaluated for non-molecular data).
Cited literature
Cavalli-Sforza LL, Edwards AWF. 1965. Analysis of human evolution. In: Geerts SJ, ed. Proceedings of the XI International Congress of Genetics, The Hague, The Netherlands, 1963. Genetics Today, vol. 3. Oxford: Pergamon Press, p. 923–933.
Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791.
Felsenstein J. 2001. The troubled growth of statistical phylogenetics. Systematic Biology 50:465–467.
Felsenstein J. 2004. Inferring Phylogenies. Sunderland, MA, U.S.A.: Sinauer Associates Inc., 664 pp. (in chapter 10, Felsenstein provides an entertaining "degression on history and philosophy" of phylogenetics and systematics).
Hennig W. 1950. Grundzüge einer Theorie der phylogenetischen Systematik. Berlin: Dt. Zentralverlag, 370 pp.
Hennig W. 1965. Phylogenetic systematics. Annual Review of Entomology 10:97–116.
Michener CD, Sokal RR. 1957. A quantitative approach to a problem in classification. Evolution 11:130–162.
Mullis KB, Faloona F. 1987. Specific synthesis of DNA in vitro via a polymerase catalyzed chain reaction. Methods in Enzymology 155:335–350.