While writing this post, the Gisaid database reported over 40,000 SARS-CoV-2 genomes (a week before it was only 32,000), which is rather a lot for a practical data analysis. There have been a few posts on the RAxML Google group about how to analyze such large datasets, and speed up the analysis:
How to run ML search and BS 100 replicates most rapidly for a 30000 taxa * 30000 bp DNA datasetIn response, Alexandros Stamatakis, the developer of RAxML, expressed the basic problem this way:
Nonetheless, the dataset has insufficient phylogenetic signal, and thus it can and should not be analyzed using some standard command line that we provide you here; but requires a more involved analysis, carefully exploring if there is sufficient signal to even represent the result as a binary/bifurcating tree, which I personally seriously doubt.As demonstrated in our current collection of recent blog posts, we also doubt this. One user, having read some of our posts, wondered whether we can't just use the NETWORK program to infer a haplotype network, instead. Typically, the answer to such a question is "Yes, but..."
So, here's a post about how I would design an experiment to get the most information out of thousands of virus genomes (see also: Inferring a tree with 12000 [or more] virus genomes).
 |
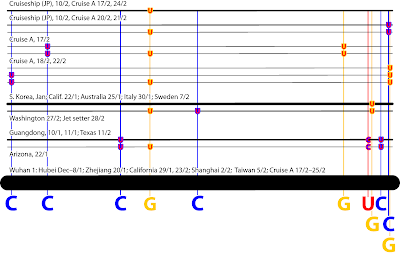
| Why trees struggle with resolving virus phylogenies and reconstructing their evolution. X, the genotype of Patient Zero (first host, not first-diagnosed host) spread into five main lineages. All splits (internodes, taxon bipartitions) in this graph are trivial, ie. one tip is seperated from all others. Thus, they, and the underlying data, cannot provide any information to infer a tree, which is a sequence of non-trivial taxon bipartitions. For instance, an outgroup (O)-defined root would require to sample the 'Source' (S), the all-ancestor, hence, defining a split O+S | X+A+B+C+D+E. All permutations of X+descendant | rest should have the same probability, leading to a 5-way split support (BS = 20, PP = 0.2). In reality, however, tree-analyses, Bayesian inference more than ML bootstrapping, may prefer one split over any other, eg. because of long-branch attraction between C and D and 'short-branch culling' of X and E. See also: Problems with the phylogeny of coronaviruses and A new SARS-CoV-2 variant. |
Start small
Having a large set of data doesn't mean that you have to analyze it all at once. Big Data does not mean that we must start with a big analysis! The reason we have over 40,000 CoV-2 genomes is simply the recent advances in DNA sequencing, and that we have effectively spread the virus globally, to provide a lot of potential samples.
The first step would thus be:
- Take one geographical region at a time, and infer its haplotype network.
We can also search the regional data for recombinants — virus may recombine, but to do so they need to come into contact, ie. be sympatric.
Go big
Once the main virus variants in each region are identified, we can filter them and then use them to infer both:
- a global haplotype network, and
- a global, bootstrapped maximum-likelihood (ML) tree.
On the other hand, the haplotype network (Median-joining or Statistical parsimony) may be biased, but it can inform us about ancestor-descendant relationships. Using the ML tree as guide, we may even be able to eliminate saturated sites or weigh them for the network inference, provided that the filtered, pruned-down dataset provides enough signal.
With the ML tree, bootstrapping analysis and haplotype networks at hand, it is easy to do things like compare the frequency of the main lineages, and assess their global distribution. This also facilitates the depiction of potential recombination, we can sub-divide the complete genome and infer trees/networks for the different bits, and then compare them.
 |
| Only based on nearly 80 CoV-2 genomes stored in gene banks by March 2020. The same can be done for any number of accessions, provided tools are used taking into account the reality of the data. The "x" indicate recombination, arrows ancestor-descendant relationships (from: Using Median networks to study SARS-CoV-2) |
Change over time
The most challenging problem for tree inference and haplotype-network inference, is the fact that virus genomes evolve steadily through time. That is, the CoV-2 data will include both the earliest variants of the virus as well as its many, diverse offspring — both ancestors and descendants are included among the (now) 40,000+ genomes. We have shown a number of examples where trees cannot handle ancestor-descendant relationships very well. Haplotype networks, on the other hand, are vulnerable to homoplasy (random convergences). So:
- Take one time-slice and establish the amount of virus divergence at that time.
- Then stack the time-slice graphs together, for a general overview.
Based on such networks, we can also easily filter the main variants for each time slice, to compile a reduced set for further explicit dating analysis, for example via the commonly used dating software BEAST (it was actually designed originally for use with virus phylogenies).
 |
| A stack of time-filtered Neighbor-nets (from: Stacking neighbour-nets: a real world example; see Stacking neighbour-nets: ancestors and descendants for an introduction) |
Networks and trees go hand-in-hand
With the analyses above, it should be straightforward to model not only the spread of the virus (as GISAID tries to do using Nextstrain) but also its evolution – global and general, local and in-depth, and linear and reticulate.
The set of reconstructions will allow for exploratory data analysis. Conflicts between trees and networks are often a first hint towards reticulate history — in the case of viruses this will be recombination. Keep in mind that deep recombinants will not necessarily create conflict in either trees (eg. decreased bootstrap support) or networks (eg. boxes), but may instead result in long terminal branches.
There may be haplotypes in the regional networks that are oddly different, or create parallel edge-bundles. Using the ML guide-tree, we can assess their relationship within the global data set — whether they show patterns diagnostic for more than one lineage or are the result of homoplasy.
Likewise, there may be branches in the ML tree with ambiguous support, which can be understood when using haplotype networks (see eg., Tree informing networks explaining trees).
Era of Big Data, and Big Error
SARS-CoV-2 data form a very special dataset, but there are parallels to other Big Data phylogenomic studies. Many of these studies produce fully resolved trees: and it is often assumed that the more data are used then the more correct is the result. Further examination is thus unnecessary (and it may be impossible, because of the amount of compiled data).
As somebody who worked at the coal-face of evolution, I have realized that the more data we have then the more complex will be the patterns we can extract from them. The risk of methodological bias will not vanish, but may even increase; and the more I then need to check which part of my data resolves which aspect of a taxonomic group's evolution.
This can mean that, rather than a single tree of 10,000 samples, it is better to infer 100 graphs that each reflects variation among 100 samples and one overall graph that includes only the main sample types. Make use of supernetworks (eg. Supernetworks and gene incongruence) and consensus networks to explore all aspects of a group's evolution. In particular, when you leave CoV-2 behind and task larger groups of coronaviruses (Hack and fish...for recombination in coronaviruses).