Category Archives: Derrick Crook
Collaborative PhD and postdoc positions available
Dr Pierre Mahe of bioMérieux in Grenoble, France, is seeking to appoint an industry-linked PhD position developing statistical methods for genome-based characterization of antimicrobial resistance and virulence genes, with a focus on the opportunistic pathogen Pseudomonas aeruginosa. The position involves a secondment here in Oxford. For more details click here or contact Pierre Mahe.
Postdoctoral Scientist in Statistical Genomics
The role is for a population geneticist or statistical geneticist to develop and apply statistical methods, including genome-wide association studies, for discovering rare and common genetic variants underlying antimicrobial resistance in Mycobacterium tuberculosis.
One third of the world's population - 2.5 billion people - are thought to be infected with tuberculosis (TB). This post offers an opportunity to work with global TB experts from five continents, statistical geneticists, clinicians, medical statisticians and software engineers; integrating statistical genetics, bioinformatics and machine learning methods with the aim of uncovering all genomic variants causing at least 1% resistance to first line anti-TB drugs.
We're looking for candidates with a PhD in genomics, evolutionary biology, statistics or a related subject. The post is full-time and fixed-term for up to 3 years initially.
The deadline for applications is noon on Friday 6th May 2016.
Posted by in Antibiotics, CRyPTIC, Derrick Crook, Genomics, GWAS, Microbiology, Mycobacterium tuberculosis, Population Genetics, Postdoc
Nature Reviews Microbiology: Within-host evolution of bacterial pathogens
 Our new review of what genomics has taught us about Within-host evolution of bacterial pathogens has been published in Nature Reviews Microbiology.
Our new review of what genomics has taught us about Within-host evolution of bacterial pathogens has been published in Nature Reviews Microbiology.Posted by in Bacteria, Derrick Crook, Evolution, Genomics, Nature Reviews Microbiology, Sarah Walker, Tim Peto, Xavier Didelot
Postdoc in antimicrobial resistance gene discovery
The role is for a population geneticist or statistical geneticist working with TB experts from five continents with the aim of uncovering all genomic variants causing at least 1% resistance to first line TB drugs. We're looking for candidates with a PhD in genomics, evolutionary biology, statistics or a related subject. The post is full-time and fixed-term for up to 3 years initially.
The deadline for applications is noon on Monday 15th February 2016.
Posted by in Derrick Crook, Genomics, GWAS, Microbiology, Mycobacterium tuberculosis, Population Genetics, Postdoc
Within-host evolution of Staphylococcus aureus during asymptomatic carriage
 Given its notoriety as one of the world's major causes of infection-related deaths, it may come as a surprise that one in three healthy adults carry the human pathogen Staphylococcus aureus in their noses without adverse effects. Indeed, most people carry the bacteria at some point in their lives. So carriage must be seen as the normal state of affairs in the human-S. aureus interaction, and by understanding this state better we can improve our understanding of why, in some people, the bacteria go on to cause life-threatening invasive disease.
Given its notoriety as one of the world's major causes of infection-related deaths, it may come as a surprise that one in three healthy adults carry the human pathogen Staphylococcus aureus in their noses without adverse effects. Indeed, most people carry the bacteria at some point in their lives. So carriage must be seen as the normal state of affairs in the human-S. aureus interaction, and by understanding this state better we can improve our understanding of why, in some people, the bacteria go on to cause life-threatening invasive disease. This month sees publication of an investigation by my colleagues and me into the evolution of S. aureus during this normal healthy carriage state. The carriers in our study harboured populations of the bacteria that were very closely related but typically not identical, implying that the bacteria had evolved within the human body. The nose appears to be a microcosm of evolution for S. aureus, showing all the different types of genetic variation known at the species level within the noses of these individual carriers. For the most part, within-host evolution of the bacteria was very conservative, but certain proteins expressed on the surface of the bacteria and toxins secreted by the bacteria showed evidence of involvement in a host-pathogen arms race.
The paper, whose lead authors include Tanya Golubchik, Liz Batty, Derrick Crook and Rory Bowden, has received coverage on the EveryONE blog and F1000. I liked Gerald Pier's conclusion, made on the post-publication peer review website: "Given that about 30% of the world's seven billion-plus humans, and an unknown number of animals, are chronically colonized with S. aureus, the tremendous opportunity provided to this organism for generating genetic variation to counteract human efforts to prevent S. aureus infections may be one of the most formidable barriers to overcome in order to develop vaccines and highly effective interventions to lessen the impact of this organism on human and animal health."
Posted by in Bacteria, Derrick Crook, Evolution, Genomics, Liz Batty, PLOS One, Rory Bowden, Staphylococcus aureus, Tanya Golubchik
Postdoctoral Positions in Pathogen Genomics
The group is studying a range of bacterial and viral pathogens including tuberculosis, Staphylococcus aureus, Clostridium difficile, HIV, norovirus and hepatitis C virus. Our research interests include within-host evolution, the genetic basis of virulence, transmission dynamics and outbreak investigation via real-time genomics.
A major translational goal of the project is to exploit the transformative effect of population genomics on bacteriology to improve routine clinical practice in public health and microbiology laboratories.
The research is supported by the UKCRC Modernising Medical Microbiology Consortium, the Health Innovation Challenge Fund, the NHS National Institute for Health Research, the Oxford Biomedical Research Centre, Institut Merieux and the Oxford Martin School, and pursued in collaboration with clinical colleagues in Leeds, Birmingham and Brighton, the Health Protection Agency and the WTSI.
The deadline for applications varies by position, between 26-28 November 2012.
For examples of recent papers see:
http://www.thelancet.com/journals/laninf/article/PIIS1473-3099%2812%2970277-3/fulltext
http://www.pnas.org/content/109/12/4550.full
http://bmjopen.bmj.com/content/2/3/e001124.full.pdf+html
http://www.nature.com/nrg/journal/v13/n9/pdf/nrg3226.pdf
http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1002874
For more information visit:
http://www.modmedmicro.ac.uk
http://www.oxfordmartin.ox.ac.uk/projects/view/127
Nature Reviews Genetics: Transforming Clinical Microbiology
You might also be interested to read a similarly themed review recently published by our friends at the University of Cambridge and Wellcome Trust Sanger Institute in PLoS Pathogens titled Routine use of microbial whole genome sequencing in diagnostic and public health microbiology.
These review articles follow hot on the heels of a pair of research articles published by our two groups: A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance in BMJ Open and Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak in the New England Journal of Medicine. The common thread is the impact of near-to-real-time whole genome sequencing on outbreak detection and other translational activities in hospitals and public health laboratories.
Post-doc Positions in Pathogen Genomics
If you are interested, please get in touch.
Posted by in Bacteria, Bioinformatics, Derrick Crook, Genomics, Hepatitis C, HIV, Lab business
PNAS paper on staphylococcal evolution during infection
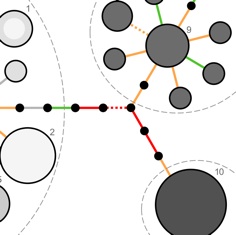 Today in PNAS Early Edition my colleagues and I have a paper published reporting the genome evolution of Staphylococcus aureus during the transition from prolonged nasal carriage to invasive disease. Since Staph. aureus, a major bacterial cause of life-threatening infections, is carried without symptoms by a quarter of healthy adults, a natural question is to ask what genetic changes - if any - accompany the transition to invasive disease. The opportunity to pursue this question arose from a detailed epidemiological investigation of asymptomatic Staph. aureus nasal carriage set up by colleagues of mine including Derrick Crook and Kyle Knox. The study has recruited over 1,000 participants in Oxfordshire since it began running in October 2008. One participant developed a bloodstream infection that was indistinguishable from the strain of Staph. aureus persistently carried in the nose for the previous 13 months. Members of the Modernising Medical Microbiology consortium, led by Derrick and Rory Bowden, sequenced the genomes of 68 bacterial colonies isolated from the nasal and blood samples from this participant, and 101 colonies from nasal samples from two other participants that did not go on to develop disease. Bernadette Young and Tanya Golubchik analyzed the genome evolution of these bacterial populations, discovering an unusual pattern in the mutations that occurred between nasal carriage and invasive disease: mutations that led to prematurely truncated proteins were significantly over-represented, including one in a gene previously associated with virulence in bacteria. To know more, read the full open access article.
Today in PNAS Early Edition my colleagues and I have a paper published reporting the genome evolution of Staphylococcus aureus during the transition from prolonged nasal carriage to invasive disease. Since Staph. aureus, a major bacterial cause of life-threatening infections, is carried without symptoms by a quarter of healthy adults, a natural question is to ask what genetic changes - if any - accompany the transition to invasive disease. The opportunity to pursue this question arose from a detailed epidemiological investigation of asymptomatic Staph. aureus nasal carriage set up by colleagues of mine including Derrick Crook and Kyle Knox. The study has recruited over 1,000 participants in Oxfordshire since it began running in October 2008. One participant developed a bloodstream infection that was indistinguishable from the strain of Staph. aureus persistently carried in the nose for the previous 13 months. Members of the Modernising Medical Microbiology consortium, led by Derrick and Rory Bowden, sequenced the genomes of 68 bacterial colonies isolated from the nasal and blood samples from this participant, and 101 colonies from nasal samples from two other participants that did not go on to develop disease. Bernadette Young and Tanya Golubchik analyzed the genome evolution of these bacterial populations, discovering an unusual pattern in the mutations that occurred between nasal carriage and invasive disease: mutations that led to prematurely truncated proteins were significantly over-represented, including one in a gene previously associated with virulence in bacteria. To know more, read the full open access article.