Author’s Note: Post was written without access only to the abstract, not the full text, of the journal article in question. Note that the argument is not with the methods or results of the research, but with how the research question has been presented.
University of Chicago Medicine & Biological Sciences tweeted the following tweet on Twitter today highlighting the work of post-doc Laure Ségurel on genetic risks for Type 2 Diabetes:
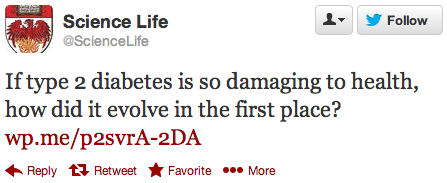
The work itself is interesting in its own right. Investigating the population genetic history of genetic markers associated with Type 2 Diabetes risk could have multiple applications, beyond the high level of intellectual interest.
The question used to frame the research, however, troubles me, because it plays to general misconceptions about the evolutionary dominance and efficiency of natural selection in humans:
Why is this deleterious disease so common, while the associated genetic variants should be removed by natural selection? -Ségurel et al (Eur J Hum Genet. 2013 Jan 23. doi: 10.1038/ejhg.2012.295)
Selection is not the only force that can drive evolution. Other options include drift, mutation, and migration. These forces are distinguished from selection by their blindness to their effects on the fitness of organisms. I use “the adaptionist paradigm” to refer to the assumption held by many, including scientists, that selection is almost always the primary driving force of evolution and that it is efficient.
Human population structure is not well suited for efficient selection due to a variety of features like not having a large effective population size (a genetics concept that can differ greatly from actual population size) and relatively long generation times. Under these conditions, it is hardly surprising that natural selection has failed to scrub all manner of deleterious genetic differences from humans, without having to postulate that what are now deleterious genetic differences were at one point “good for us”.
At the same time, I hardly blame the authors for taking this approach to present their work [UPDATE 9 Mar 2013: Underlined text added to clarify that "approach" does not refer to how the research was conducted-see comment thread]. Their research directly addresses the “thrifty” genotype hypothesis (insulin resistance was beneficial for hunter-gatherers) for Type 2 Diabetes genetic risk factors, which still has a lot of traction. It is an adaptionist hypothesis addressing a problem emerging from the adaptionist paradigm. That this problem is not well supported by evolutionary theory does not change the fact that it is relevant to the field of research.
There is also an issue of the genetic differences and this one requires paying close attention to the jargon. In particular, we need to distinguish between the words associated and causal:
The ‘thrifty genotype’ hypothesis proposed that the causal genetic variants were advantageous and selected for during the majority of human evolution. It remains, however, unclear whether genetic data support this scenario. In this study, we characterized patterns of selection at 10 variants associated with type 2 diabetes… -Ségurel et al (Eur J Hum Genet. 2013 Jan 23. doi: 10.1038/ejhg.2012.295) [Emphasis mine]
Causal genetic differences are the ones that actually cause the increased risk. These are the locations where having one base pair instead of another means you have a different likelihood to get the disease. Associated genetic differences are relatively common genetic variations in the human population that can be linked statistically to risk. Associated differences are markers for causal differences, because they are usually, but not always inherited together. It’s like the golden arches sign for McDonald’s. The sign is not the McDonald’s, but it is almost always right next to the McDonald’s; and it strongly suggests the nearby building will not be a Burger King.
We tend to use associated differences (thus, genome-wide association studies) because the large populations necessary for these studies need common variants and working with a known set of differences is much easier technologically (ie, much easier to genotype people).
This means that the researchers are, at some level, using a proxy for the evolutionary history of the causal differences. This can introduce multiple confounding issues, for example the causal mutation entering the population after the associated difference making it a poorer quality marker. In such a case, natural selection could act to remove the causal difference, but leave the associated marker – though, in that case, the marker would no longer be associated with risk [UPDATE 19:53 - a version issue caused this sentence to be truncated when posted].
Most genetic markers only contribute marginally, but statistically significantly to a person’s individual risk. Their individual effect on individual fitness may not particularly large. In addition, Type 2 Diabetes frequently manifests at an age when people have already reproduced. Pile on top of that the fact that the human population structure (this pretty much goes for all large animals) does not have the characteristics necessary for the efficient operation of natural selection.
For my money, the better motivating question and the question the research really asks is:
Is selection acting on genetic differences associated with Type 2 Diabetes & is it favoring the risky or protective differences?
What they found was that selection is acting on genetic differences associated with Type 2 Diabetes risk and is favoring the protective variants. They also found that these results did not appear to depend on the lifestyle of the populations studied, refuting the ”thrifty” genotype hypothesis. Selection seems to be favoring the genetic differences that protect against Type 2 Diabetes and it does not appear to be related to the lifestyle of the populations being studied. In doing so, it answers my questions, which I happen to believe are the “right” questions, but also perpetuates misconceptions about evolution.
I do understand why the researchers took this approach to present their work [UPDATE 9 Mar 2013: Underlined text added to clarify that "approach" does not refer to how the research was conducted-see comment thread]. The study is framed in a way that is relevant to the field, if not necessarily the science, which is kind of a shame, when you think about it.
*This post is also a reminder to me to make sure you check what the researchers said about the topic, instead of jumping to the conclusion that the University press officer got carried away trying to explain why a study is “interesting”. In this case, the public article reflects what the researchers wrote in their peer-reviewed, subscription access journal article.